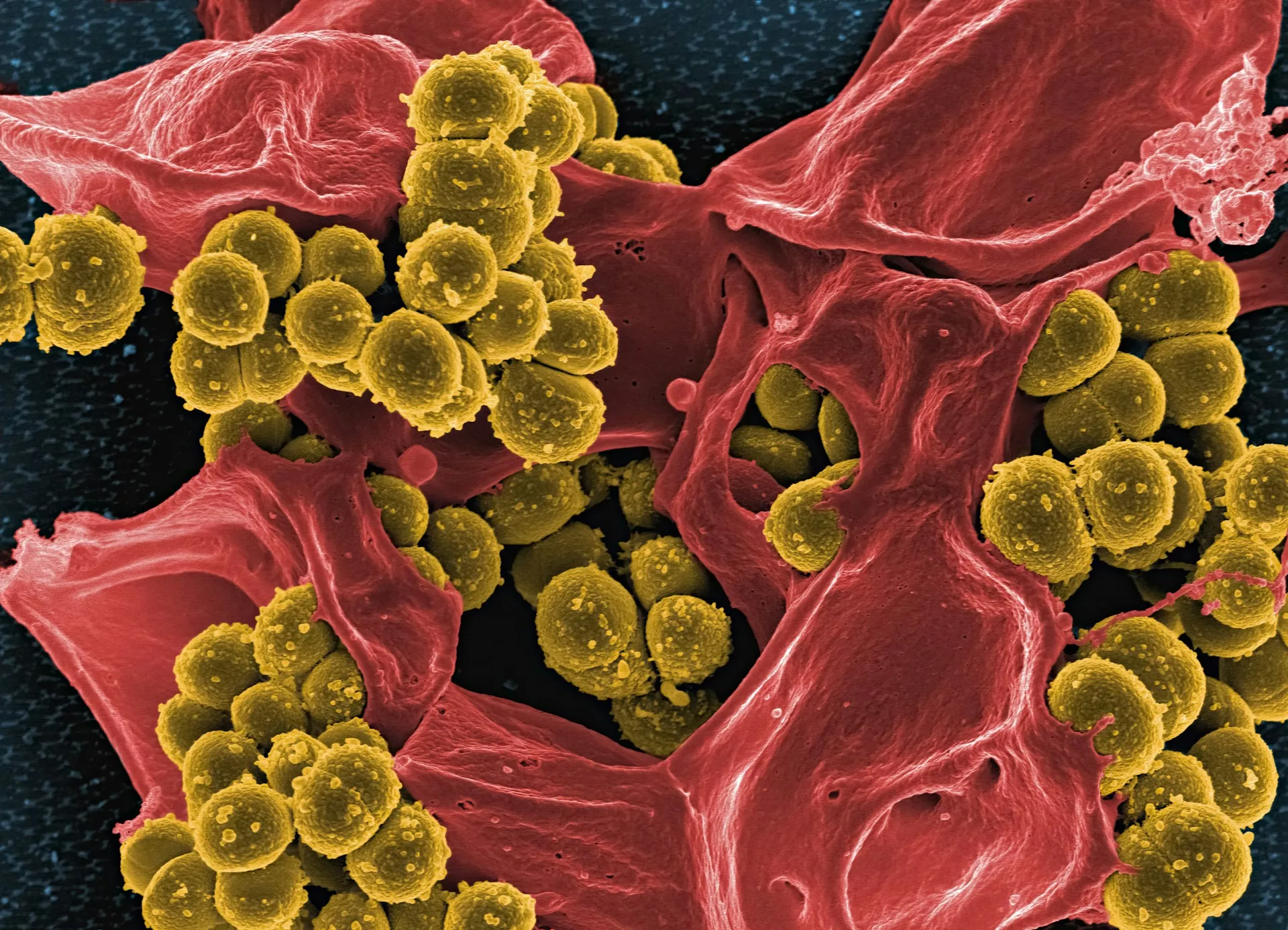
Our extensive use of antibiotics has led to resistance evolving, MRSA is one of the deadliest resistant pathogens. New findings using AI may help overcome this issue. Photo credit: National Institute of Allergy and Infectious Diseases via Unsplash
This [MRSA antibiotics] could be the first new class of antibiotics introduced in decades…
Scientists have discovered a novel structural class of antibiotic compounds effective against methicillin-resistant Staphylococcus aureus (MRSA), the deadliest drug-resistant pathogen worldwide. This could be the first new class of antibiotics introduced in decades — and it is all thanks to artificial intelligence.
This groundbreaking development comes from a team based at the Broad Institute of MIT and Harvard, led by Felix Wong and Erica Zheng with help from researchers in Germany. Their work forms a part of the Antibiotics-AI Project at the Collins Lab in MIT, a revolutionary project aiming to uncover seven new antibiotic classes in seven years by harnessing the power of AI. The project boasts previous successes that include the discovery of halicin and abaucin, antibiotics effective against highly resistant bacterial strains such as Mycobacterium tuberculosis and Acinetobacter baumannii. However, this time the researchers turned their focus to MRSA—a notoriously resistant ‘superbug’ responsible for over 100,000 deaths in 2019.
In their quest for novel antibiotic candidates the team used deep learning, a form of machine learning that mimics the cognitive processes of the human brain. This cutting-edge technique involves generating models that can make predictions by learning from information (known as ‘training data’). In this study, the models were trained to predict both antibiotic activity against MRSA and toxicity to human liver, muscle, and lung cells using a dataset of 39,000 compounds with known properties. Once satisfied with the models’ accuracy, the researchers used them to scour an enormous library of over 12 million compounds, scoring each on its predicted efficacy and safety. By analysing these scores, the team could then easily identify promising antibiotic candidates—saving them valuable time compared to sifting through the compounds themselves.
In their quest for novel antibiotic candidates the team used deep learning, a form of machine learning that mimics the cognitive processes of the human brain.
Amazingly, the models successfully identified two new compounds sharing a common substructure previously unseen in antibiotics. Upon testing, both exhibited remarkable efficacy against MRSA, outperforming conventional antibiotics such as ampicillin and ciprofloxacin. Further investigation also demonstrated that the compounds could reduce bacteria levels in a live mouse infected with MRSA and proved their non-toxicity to human skin. What’s more, these potent compounds were also effective against a further 40 different resistant bacterial strains, demonstrating their broad-spectrum potential. Amongst these included vancomycin-resistant Enterococcus (VRE), which features alongside MRSA on the WHO priority list of resistant pathogens for which new antibiotics are urgently needed.
Wong and his colleagues highlight that a key feature of their deep learning approach is that it is explainable, meaning the antibiotic activity of compounds can be traced back to specific chemical substructures within them. In this particular case, the compounds’ shared substructure appeared to work by disrupting pH gradients across the bacterial cell membrane. Rather interestingly, antibiotic candidates with this mechanism of action have previously been discarded for not being selective enough — whilst these only targeted bacteria with a specific cell wall structure, known collectively as ‘gram-positive’ strains.
Findings such as these could not be more critical amidst a three-decade long ‘drought’ in antibiotic discovery and alarmingly high rates of resistance. With 2024 set to be another enormous year for AI-powered drug discovery, hundreds of new antibiotics could be just around the corner.