Ten years ago, the completion of the Human Genome Project was a milestone in the history of science. The race to be the first team to decode an entire human genome ultimately ended with the two largest groups collaborating and announcing the success of the project in 2001. This was not only a scientific achievement, but also an ideological one, with the project spanning 15 years and fuelling an era of technological advances in DNA sequencing which continues to this day.
One such advance was announced less than a month ago, when a UK research group led by Matt Loose at Nottingham University announced they had sequenced a strand of DNA 10,000 times longer than the usual standard length, using a technology known as nanopore sequencing.
A UK research group led by Matt Loose at Nottingham University announced they had sequenced a strand of DNA 10,000 times longer than the usual standard length
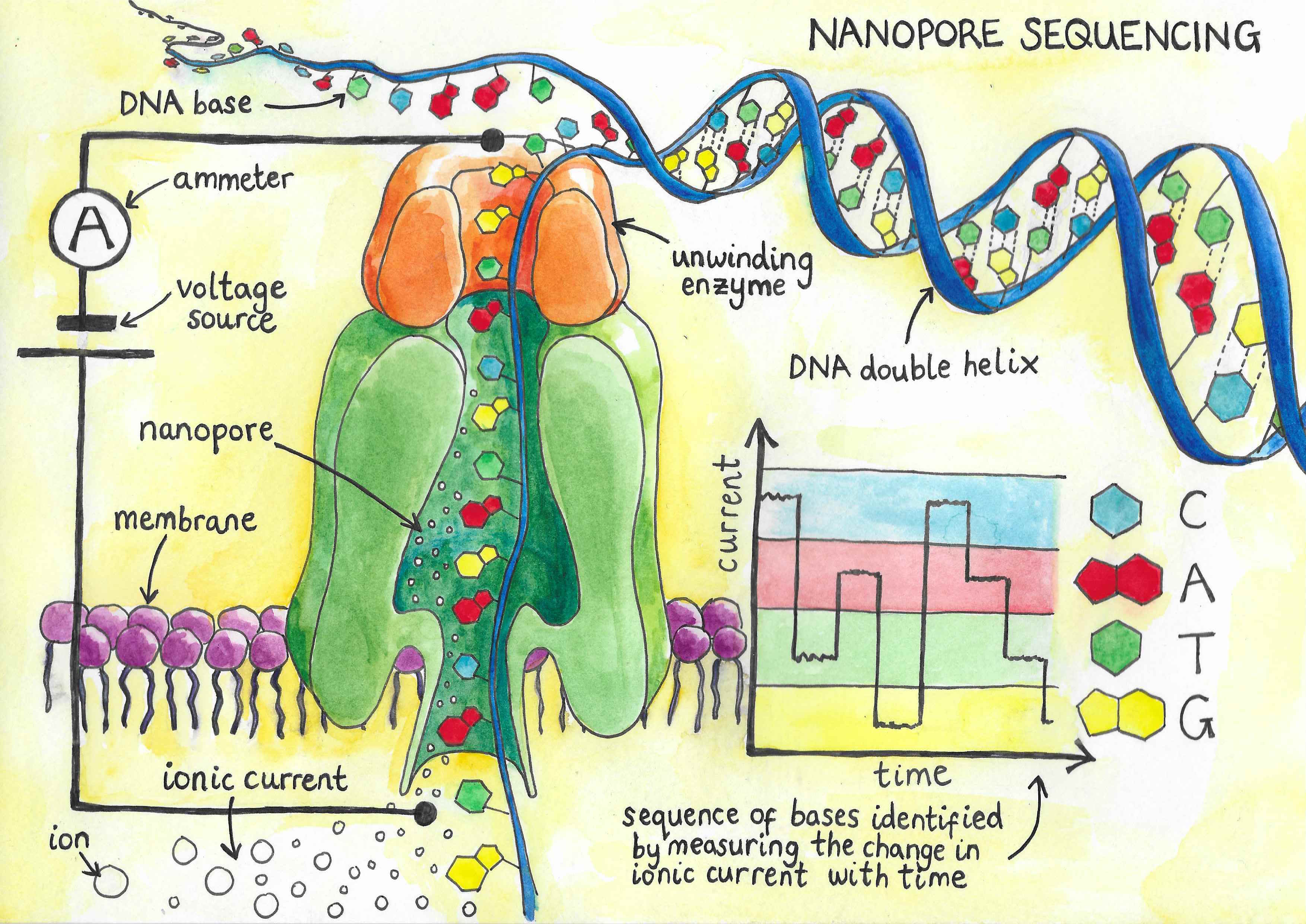
Sequencing DNA allows us to decode the information stored inside a cell and then better understand how it behaves. Until now, DNA sequencing technology has required the DNA to be chopped up into small sections that are then sequenced and reassembled by a computer to generate a copy of the genome called an assembly. This process is similar to solving a jigsaw puzzle – overlapping ends of the short fragments are compared, and the sequence is assembled by comparing the matching regions. The data can be hard to assemble into a complete sequence, especially in regions that look similar to other parts of the genome or where there are many repeats of short sequences. This in turn makes it difficult to determine how many repeats are present or which sequence belongs to which region of the genome, leading to gaps in the assembly. Because of this, there is an incentive towards developing long-read sequencing techniques that allow longer continuous sequences to be decoded and to be able to sequence such a long strand of DNA in a single run is a major breakthrough.
Nanopore sequencing, developed in 2008, works by using an electric field to pull DNA towards a positive charge. In order to reach the positive charge, the DNA has to travel through a narrow pore in a membrane. This pore has a diameter of just 1.5 nm at its narrowest point, meaning that only single strands of DNA can pass through. As the strand goes through the pore, the bases – which reflect the letters in the DNA sequence – block the opening. This causes minute changes in the electrical conductance of the membrane, which can be recorded. As each base has different properties based on its physical structure, a unique profile of duration and magnitude of the pore blockage for each base can be identified and used to determine the identity of the base. In this way, the sequence of the strand passing through the pore can be identified, with the length of the read depending only on the successful preparation of this fragment. The technique is extremely fast, so that if the entire human genome were encoded in a single DNA fragment, it could be decoded in just 20 hours. Additionally, the cost of sequencing is significantly lower than existing techniques, as fewer expensive chemicals and specialist materials are needed.
[Nanopore sequencing] is extremely fast, so that if the entire human genome were encoded in a single DNA fragment, it could be decoded in just 20 hours.
A significant amount of work on nanopore sequencing has been done in Oxford. The first pore that could distinguish all four bases of DNA was identified by a research group at the Department of Chemistry, who were also the first to successfully use nanopore technology to sequence a whole strand of DNA. The technique is currently being developed for commercial use by Oxford Nanopore Technologies, which has released several devices for nanopore sequencing. One of these systems, the MinION, consists of a small, disposable sequencing chip that contains the nanopores, required fluidics, and electronic sensors, and is approximately the size of a small cell phone. All that is needed to use it is the prepared DNA fragment and a laptop with a USB port to plug it into. The portability of devices like the MinION is one of the aspects that gives nanopore sequencing such great potential: sequencing can now be done anywhere, in real-time, without access to a laboratory and the large machines that were previously needed to sequence DNA. This allows the MinION to perform rapid and mobile DNA sequencing with ease.
These advantages have not escaped notice, and the MinION has begun to be used to acquire information rapidly in clinically relevant situations both in remote and urban areas. The device was used to identify Salmonella strains following an outbreak in a British hospital, identifying the strain far faster than, and just as accurately as, other major sequencing techniques. Perhaps most excitingly though, the MinION allows sequencing to occur in areas where little laboratory infrastructure is available by eliminating the need for expensive and bulky equipment. In the recent West African Ebola epidemic, researchers used the devices to sequence the viral genomes from 14 patients within only 12 days. The viral genomes can then be used for research purposes, to track the evolution and spread of the virus and, importantly, to diagnose Ebola and other endemic diseases, such as Chikungunya Virus or Hepatitis C.
In the recent West African Ebola epidemic, researchers used the devices to sequence the viral genomes from 14 patients within only 12 days.
The record-setting DNA read recently obtained by the Nottingham group is part of the trend towards faster, cheaper long-read sequencing technologies. These developments will aid the identification of pathogens, diagnosis of infections, and enable tracking of disease outbreaks in real-time. It’s not all about saving lives though. Scientists are a competitive bunch and a big driver of these innovations is the friendly competition that occurs between research groups. This is embodied by the recent creation of a trophy that is allocated to the group that holds the record for the longest read. For now, the trophy sits in pride of place in Nottingham, where it will remain until the next advance in long-read sequencing whisks it away.
A big driver of these innovations is the friendly competition that occurs between research groups.
Artwork by Penelope Streatfeild