Dr. Victor Padilla-Sanchez, PhD, CC BY-SA 4.0, via Wikimedia Commons
While in recent years the CRISPR system has gained fame for its use in gene editing, its origins date back to the immune systems of bacteria and archaea defending against viral attack.
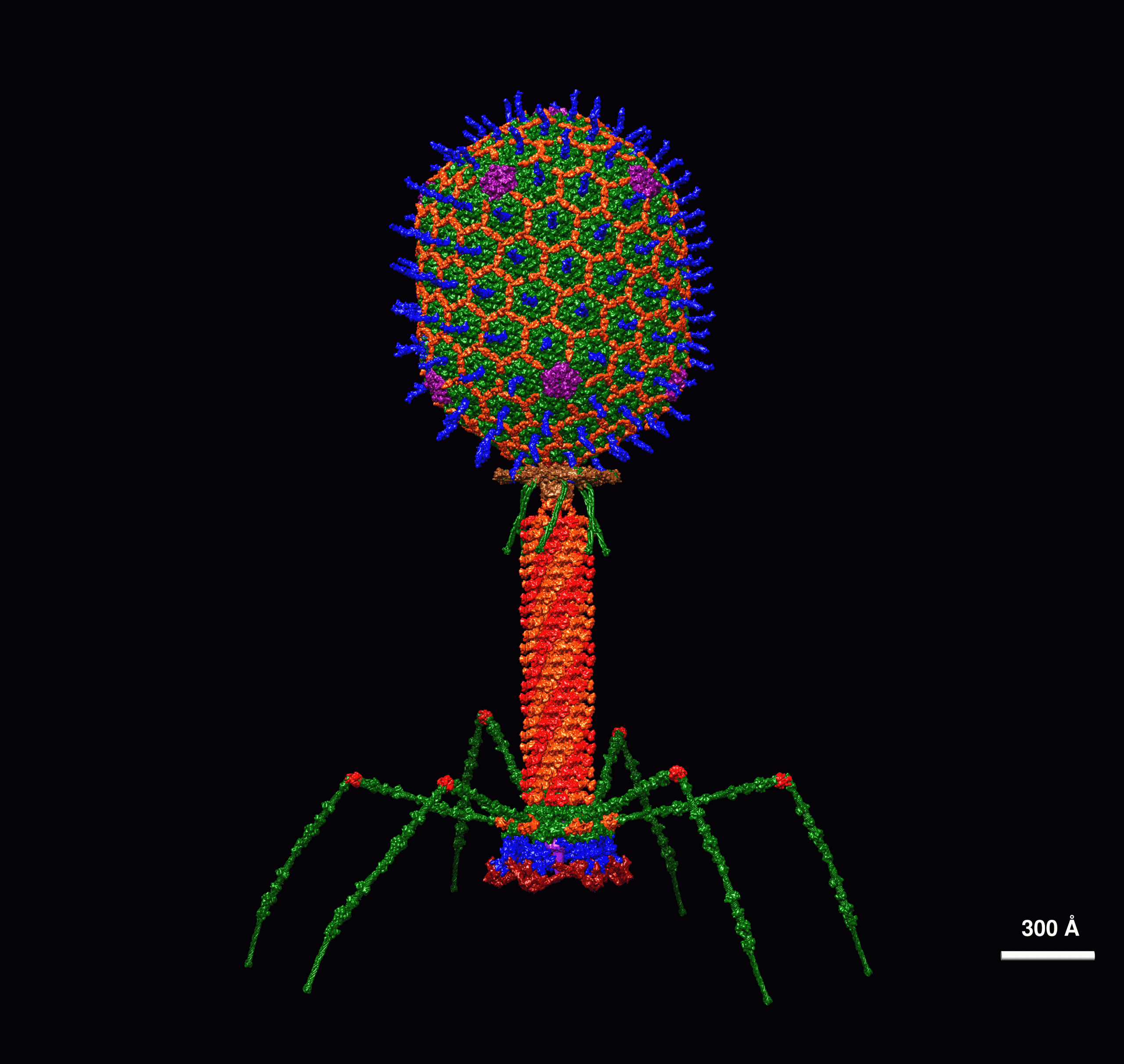
A new study has unveiled that these invading viruses—also known as bacteriophages (or “phages,” for short)—in fact have the ability to steal the CRISPR system and weaponise it against other competing phages, defeating their rivals to keep the bacterial hosts for themselves.
Standing for ‘Clustered Regularly Interspaced Short Palindromic Repeats,’ the CRISPR molecular system in partnership with CRISPR-associated proteins (often shortened to “Cas”) acts like a pair of scissors to cleave parts of invading phages and store them within the bacterial genome.
This creates a library of viral DNA fragments, like a memory bank, so that reinfection with the same virus can be recognised by Cas and the attack halted.
These Cas proteins are numerous forms of enzymes which recognise RNA created by the CRISPR template and can then go on to destroy the phage. A CRISPR-Cas system has been suggested previously for viral genomes but in isolated cases, prompting a much more thorough investigation into whether phages are stealing this machinery from their hosts.
When Professor Jennifer Doudna (University of California, Berkeley) and Professor Emmanuelle Charpentier (Max Planck Unit for the Science of Pathogens) published their 2012 paper on adapting CRISPR as a new gene-editing tool, they set the precedent for gene-editing in the modern age (although this was not without patent disputes).
This ground-breaking technique allowed for extremely precise gene-editing, and unsurprisingly lead to a Nobel Prize in Chemistry for both women in 2020.
But Doudna’s work did not stop there. Along with the microbiologist Professor Jillian Banfield (UC Berkeley), their team has recently completed an exploration of phage DNA by using a technique called metagenomics, where analysis is performed directly within an environmental sample; in this case, of human, animal, aquatic, and soil microbiomes, where there is bacteria in abundance for phage invasion.
Their report in Cell reveals the presence of rare CRISPR-Cas systems in 0.4% of the phages studied; prevalence in sampled bacteria is approximately 100-fold higher at 40%, and even more with 85% of archaea containing CRISPR-Cas systems.
Why does this matter? The percentage of phages exhibiting these systems may sound small, but the significance of these findings is in their diversity rather than only number.
Over 6000 types of phages were found to contain CRISPR DNA spanning all six CRISPR types, which are associated with different properties. These include cutting key RNA sequences, or down-regulating the abortive infection pathway, which allows host cells to self-destruct and prevents the phage from spreading to nearby cells.
The structure of the CRISPR-Cas systems varied, with some showing surprising compactness and others missing key components, indicating a high diversity for these phage systems.
Doudna, Banfield, and colleagues also delved into the properties of the viral Cas enzymes, presenting the small Casλ (lambda) as a particularly interesting example. A notable finding was the small size of phage CRISPR-Cas proteins, which in the case of Casλ was able to edit the genomes of lab-grown human, wheat, and cress cells (Arabidopsis thaliana) without sacrificing efficacy.
A more compact enzyme is easier to deliver into cells, compared to the Cas9 used currently which is much larger, making these viral enzymes a potential tool for genome editing.
This is a promising discovery in our as-yet-incomplete understanding of CRISPR technology. Future research will be needed to uncover exactly how these phage CRISPR systems interact with their hosts, and whether scientists can adapt this for gene-editing. But this is certainly a promising start.