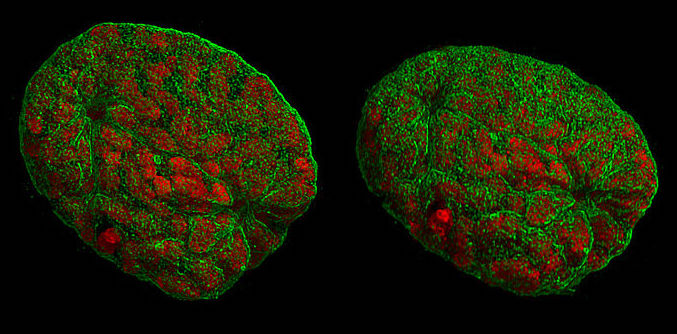
A representation of mouse nuclei during prophase, taken with 3D-structured illumination microscopy. Like other types of super-resolution microscopy, this technique allows visualisation of structures smaller than light’s diffraction limit. Lothar Schermelleh, CC BY-SA 3.0 , via Wikimedia Commons.
This article originally appeared in the Oxford Scientist‘s Michaelmas Term 2022 Print edition, Barriers, under the title ‘Super-resolution microscopy: shining light beneath the optical barrier’.
If you shoot somebody a glance, what do you shoot? The metaphor, obviously symbolic today, is not so far-fetched. Euclid and Ptolemy imagined that people could see with rays that shoot from their eyes. Whilst this theory is deeply flawed, as light enters the eye from outside, the geometrical conclusions have proved useful in explaining mirrors and lenses. The concept of light as rays or small particles persisted and informed the understanding of optics throughout the European and Islamic Middle Ages and into early modern times. The first microscopes appeared in the early 17th century, and opened up a microcosm of biological structure and movement that culminated in the discovery of cells. Geometric optics—characterising light as rays—was still the preferred explanation for how magnification works.
The utility of a theory, however, is no guarantee for truth. Through the 19th century, but based on earlier work of Christiaan Huygens, it emerged that rays of light only represent an approximation of the truth. Many phenomena are better explained by treating light as waves, an example of which is diffraction. When waves encounter an obstacle of a similar size to their own oscillating length, they bend around the object. This places a crucial theoretical limitation on the resolution of microscopes, known as Abbe’s diffraction limit. Using visible light, details below around 250 nanometres cannot be resolved, which is sufficient for researchers to observe cells, but not to distinguish many of their components.
Despite significant advances in microscope design up to the diffraction limit, throughout the 20th century biologists needed to proceed with alternative techniques to examine smaller features. As narrower waves allow for higher resolution, one possibility was to look beyond light. Fast-moving electrons behave like waves of much higher energy, and so electron microscopy can resolve structure down to single atoms. The powerful impact of electrons, however, is extremely damaging to biological matter. Therefore, the sample needs to be stabilised by staining with heavy metals or cooling with liquid-nitrogen. Either way, sample preparation is a limiting factor and live cell imaging becomes impossible.
Methods to see sub-diffraction structures in visible light were only developed at the turn of the 21st century, and were rewarded with the 2014 Nobel Prize in Chemistry. These solutions rely on the physical process of fluorescence. When a nightclub turns on the UV lamps, certain materials respond by emitting light in the visible spectrum. The responsible chemical groups can be of great use to science if they are attached to a molecule of interest, acting as a bright marker in front of a dark background. Beyond revealing the subcellular organisation of DNA or proteins, fluorescence aids resolution because it can be switched on and off.
One variant of super-resolution microscopy uses a beam of light to temporarily deactivate fluorescence, save for a tiny spot. When a second beam hits the sample, only molecules that have remained active light up, which effectively increases the resolution. Another technique is based on observing individual fluorescent molecules. While a single look only allows for accuracy in the diffraction limit, through many measurements the true position can be approximated. This does not yet increase resolution, because it still fails to distinguish between two molecules that are close together. Nonetheless, when fluorescence is activated and deactivated at random, only one of two neighbouring dots will be active in a given measurement, and a computer can reconstruct the complete picture.
Over the last decade, these approaches have swiftly moved from proof-of-concept to widespread use. The result is a stream of unprecedented views of cellular features, such as the cell’s skeleton. Yet, with the diffraction barrier broken, some challenges have merely evolved. For instance, sample preparation is often more advanced and post-processing more complex. Additionally, the required high intensities of light can degrade the sample, and for now, the technique is still expensive.
This illustrates not only a shift in what we can see, but also how we see. Euclid and Ptolemy may have been wrong in suggesting active visual rays. Nevertheless, microscopic vision is becoming a more hands-on and active process, where careful design of markers and experimental protocols drives sub-diffraction exploration. Barriers can be removed more or less suddenly, but the study of what lies beneath is an interdisciplinary voyage that we are just setting out on.