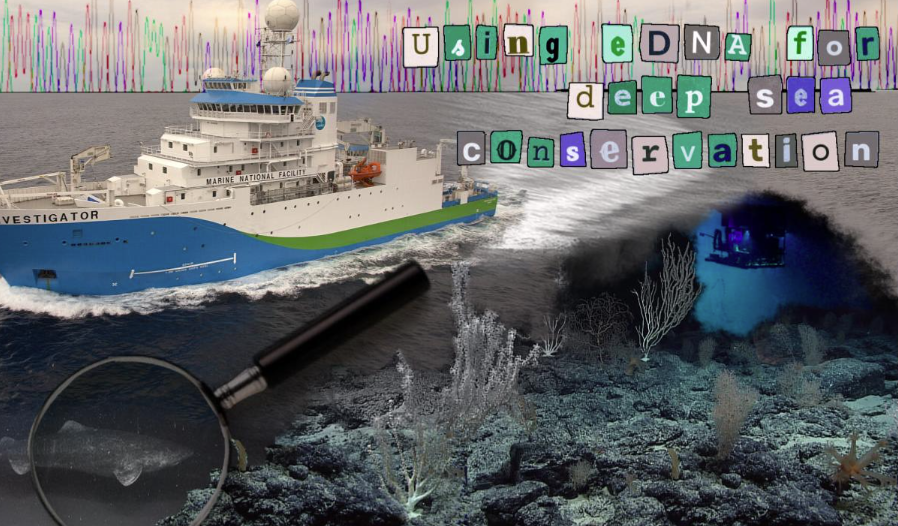
New genetic tools are helping scientists uncover hidden life in the deep ocean, reshaping how these remote ecosystems can be protected. Photo credit: Image by CSIRO, CC BY 3.0. Collage by Lucy Back via the Oxford Scientist
This article is co-authored by Lucy Back and Sofia Raffio-Curd.
In conservation, it is generally useful to know where to find the thing you’re trying to conserve. This is an easy enough task for more familiar species, but no matter how all-terrain your vehicle is, it’s notoriously difficult to hunt something down at the bottom of the ocean. Nonetheless, the deep sea is home to some of the most fascinating species on Earth—species that could soon be under threat by new industries such as deep-sea mining. So how does one go about conserving some of the least hospitable habitats on the planet? Some researchers believe that eDNA is the answer.
Environmental DNA, or eDNA, is DNA that is shed by living things as they go about their daily lives. It leaves an invisible trace behind an organism and gives us insights into the biodiversity of an area of interest. eDNA samples have been used with great success for studying the range of land organisms, especially those that are difficult to spot in person, such as the European mink. It has also been used in studies on single-celled marine organisms, but only recently have there been any studies on the eDNA of marine vertebrates. In industry, eDNA is often used for biodiversity surveys and impact assessments as it is low-cost and easy to use.
Because sampling can be automated, eDNA can be used to collect data from inaccessible environments such as the deep sea. Research vessels often drag camera systems behind them to image the ocean floor and associated wildlife (Nautilus Live, for example). Adding an eDNA collection system to this array allows samples to be taken along a transect, as well as providing a second data source for comparison. Organisations such as OceanOmics, an Australian not-for-profit, are utilising eDNA to better understand marine communities. In collaboration with the University of Western Australia, they aim to create a comprehensive database of marine vertebrate genomes in the Indian ocean, as well as studying deep sea environments which remain poorly understood.

eDNA from marine organisms can be sampled using modified equipment already in use on deep sea research vessels. Illustration by Lucy Back.
eDNA data comes with some caveats. Terrestrial studies take their samples from stationary objects and surfaces, but marine studies take samples from seawater. Due to the constant motion of water, researchers cannot be sure that eDNA in that sample is from that location. In general, DNA degrades within days or even hours in warm weather, so it can’t drift far before the signal is lost. But what about DNA in colder, deeper waters? Studies suggest that DNA in water under ten degrees Celsius could last for weeks or even longer, and may have drifted for tens of kilometres before sampling. Furthermore, the deep sea could act as a reservoir of sunken eDNA from shallower waters, complicating findings even further. Together, these factors make interpretation of deep sea eDNA data challenging.

As shown in this cross-section, water temperature drops off quickly in deep sea environments, which slows DNA degradation. Data courtesy of Amentum Scientific.
Fortunately, this is one of many cases where big data can save the day. Thanks to easy-access oceanographic datasets (such as those available via Amentum Scientific), it is possible to make estimates about the origin time and location of eDNA. Using records of water temperature, researchers can predict how long DNA may have been drifting for and use this to assess the reliability of their samples. Furthermore, it is possible to use ocean currents to backtrack and estimate the origin region of a sample. Potentially, you could even use ocean forecasts to take multiple samples over time from the same origin location, regardless of moving currents. Taking currents and temperatures into account is particularly important when using highly sensitive species-specific sampling methods, which can pick up even small traces of DNA, or when searching for a species with a small range where accuracy matters.

Sea-surface temperature heatmap with overlaid ocean currents off the east coast of Australia, indicating how temperature and currents could affect the drift of eDNA over time. Data courtesy of Amentum Scientific.
eDNA could be the future of marine conservation research. Together with a more comprehensive understanding of ocean conditions, it could be used to reliably capture community composition and species range for some of the least-studied and most vulnerable regions on Earth, leading to better-informed conservation solutions which benefit us all.
This article was created as part of a micro-internship with Amentum Scientific via the Oxford University Careers Service.